OpenADMET Consortium
Next generation ADMET modelling for therapeutic development
Small molecule therapies represent a significant portion of FDA-approved drugs, yet their development is often hindered by the challenge of balancing on-target activity with desirable pharmacokinetic (PK) properties.
Pharmacokinetic properties, which encompass absorption, distribution, metabolism, and excretion (ADME), determine a drug’s fate within the body and are crucial for its efficacy and safety. Currently, the optimization of these properties is often addressed late in the drug discovery process, leading to costly late-stage failures.
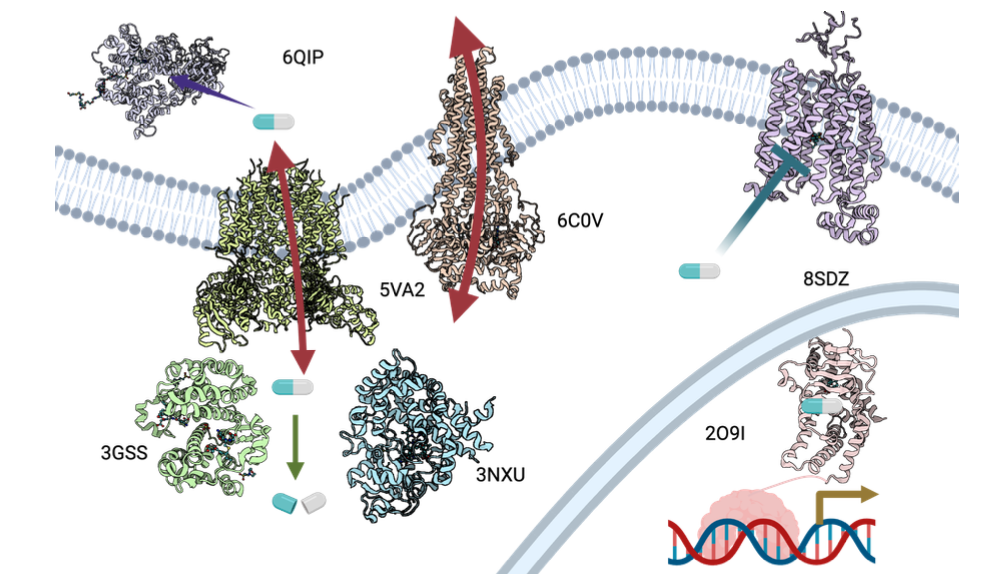
The OpenADMET project seeks to overcome this limitation by proactively characterizing the chemical space accessible to ADMET-associated proteins (“anti-targets”). By applying recent advances in experimental and computational techniques, a comprehensive open library of experimental and structural datasets will be generated.
The precompetitive resources developed by OpenADMET will provide valuable insights into the binding properties of “anti-targets” and empower researchers to develop predictive AI models for pharmacokinetic optimization. This shift towards a more proactive and data-driven approach promises to significantly expedite drug discovery by streamlining lead optimization, mitigating late-stage attrition, and ultimately accelerating the delivery of new therapies to patients.
OpenADMET is funded through an ARPA-H grant: “AVOID-OME: Structurally enabling the “avoid-ome” to accelerate drug discovery”. Read more on the AVOID-OME concept here and why we think turning the tools of modern drug discovery on the AVOID-OME can create meaningfull progress in drug discovery.